Correlation Between Mendelian and Complex Disease
Disrupted genes in Mendelian syndromes have large effects on downstream targets, contributing to the multitude of syndromic features. One or more of these gene targets may affect the risk of common disease. Recent studies have demonstrated a combinatorial effect of Mendelian syndromes on risk of common disease, but until recently, with the advent of high throughput sequencing, we have not had the ability to systematically detect these interactions within an individual patient. While the relationship between Mendelian syndromes and common diseases acts through multiple layers of cellular regulation, we take a focused approach to study interactions at the level of chromatin structure (Hi-C, ChIP-seq and ATAC-seq) and transcriptional regulation. Understanding the joint influence of monogenic mutations and common disease variants on disease phenotypes allows us to unravel the underlying biological processes contributing to human disease.
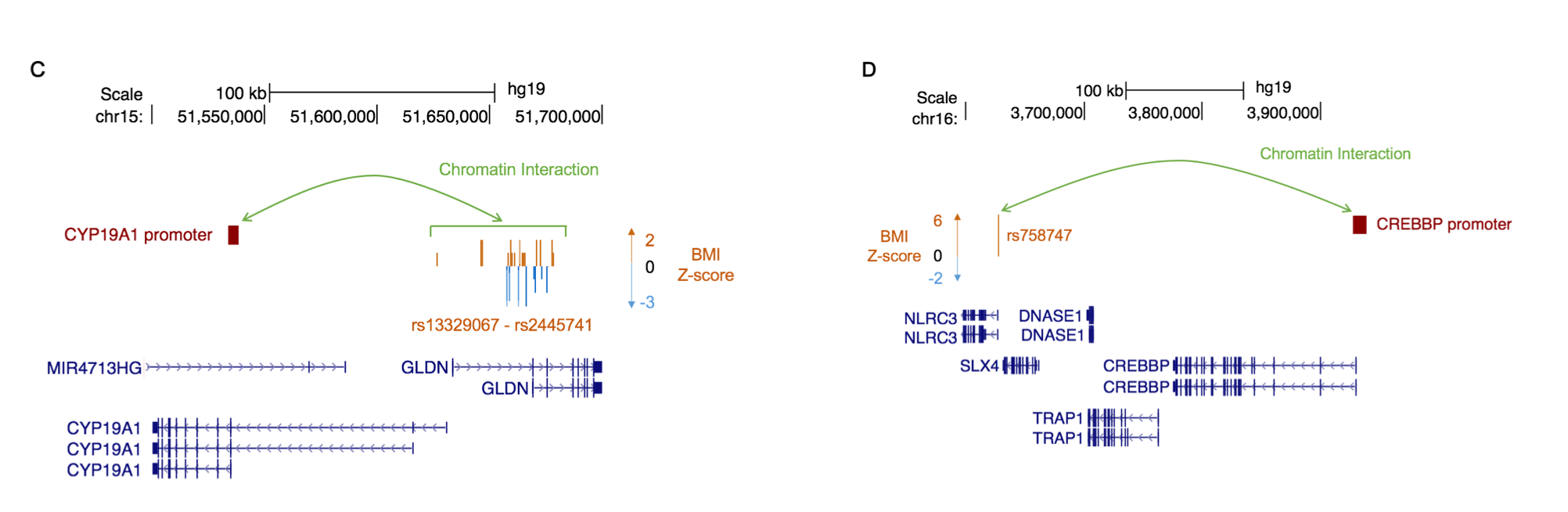
The SNP effect size in a GWAS study is larger when the SNP is localized near a phenotype-matched Mendelian gene.
Our more recent work has focused on quantifying the contribution of phenotype-specific mendelian genes to SNP-effect sizes from genome wide association studies (Freund MK, et al AJHG 2019). We found that when explored the effect of phenotype-matched Mendelian Disorder genes, the nearby GWAS signal had a larger effect. This suggested to us that non-coding variation that can modulate expression of Mendelian disease genes affect the same clinical phenotypes as those associated with the Mendelian disease, but with a more subtle effect.
To full explore these questions, we are interested in leveraging CRISPR based screen to test the effects of non-coding interactions on cell-type specific long-range interactions (see below).
Long range interactions in pre-adipocyte promoter Hi-C data demonstrate that specific GWAS variants have long-range interactions with phenotype-matched genes for BMI.